How do we identify cluster members?
Members of the Little Scottish Cluster are identified either by their haplotype (STR results) or by their haplogroup (SNP results). In terms of SNP results, you need to test positive for either of the SNPs S424 or S190. In terms of STR results, you need to have certain values for specific characteristic markers. The more markers you test, the more certain you can be of your membership.
Characteristic STR Markers
The problem of identifying STR clusters and their members can be challenging. Clusters are comprised of men with similar haplotypes. There is no precise way to specify how similar a haplotype has to be to the other members in order for that man to be considered a member of the cluster. Most approaches will require the candidate's haplotype to be within a certain genetic distance of some reference haplotype, or perhaps for it to share certain unusual marker values with the other cluster members. Regardless the approach, there will always be borderline cases where judgement calls need to be made. Only true genetic clades defined in terms of SNPs can be unambiguously defined.
Fortunately the Little Scottish Cluster is fairly isolated from a haplotype perspective, and most membership determinations are fairly obvious. There has been one very important case where membership was not very clear at all. The man had the surname Reddin, and his DNA now has a unique position within the structure of the LSC. More can be learned about him in the SNPs and Haplogroups section of Y-RESULTS.
For all the rest of us, membership is determined by comparing a candidate's haplotype against the modal haplotype of the cluster. Most men within the R1b haplogroup have very similar marker values on the majority of their markers, and only differ on a fraction of them. To focus on the signal, and elimate the background, a candidate needs only compare his marker values to the modal haplotype values that are different from the average R1b man. Often these are called the characteristic markers, and they are the "peculiarities" that have been passed down from the cluster's founder. For the Little Scottish Cluster, the complete list of characteristic markers are given in the table below:
| Marker | Typical R1b-L21 Value | Typical LSC Value(s) | FTDNA Test |
|---|---|---|---|
| DYS391 | 11 | 10 | 12 Marker |
| DYS458 | 17 | 18 | 25 Marker |
| DYS449 | 29 | 30 | 25 Marker |
| DYS464 | 15-15-17-17 | 13-15-17-X | 25 Marker |
| GATAH4 | 11 | 10 | 37 Marker |
| DYS570 | 17 | 18 | 37 Marker |
| DYS590 | 8 | 9 | 67 Marker |
| DYS413 | 23-23 | 22-23 | 67 Marker |
| DYS444 | 12 | 11 | 67 Marker |
| DYS446 | 13 | 14 | 67 Marker |
| DYS710 | 35 | 31 | 111 Marker |
| DYS495 | 16 | 15 | 111 Marker |
| DYS717 | 19 | 20 | 111 Marker |
| DYS505 | 12 | 13 | 111 Marker |
| DYS533 | 12 | 13 | 111 Marker |
| DYS715 | 24 | 23 | 111 Marker |
The more markers a candidate has tested, the more confident you can be that the candidate does or does not belong to the Little Scottish Cluster. Both the values DYS391 = 10, and DYS391 = 11 are very common amoung R1b men, and so no determination can be made at all from just 12 markers results. DYS464 in the 25 marker set of results is the first real hint of membership in the LSC. The value of 13 for DYS464a is uncommon within R1b, but is not exclusive to the LSC. There is another cluster of men called Irish Type III which also has DYS464a = 13, and can easily be confused with the LSC if only 25 markers are compared.
At 37 markers a couple more distinguish characteristics are tested, and one can generally be confidence of membership. The most definitive matches require at least the 67-marker test from FTDNA. This test includes the marker DYS590 for which men in the Little Scottish Cluster invariably have the value 9, while most other men in R1b have the value 8. I always encourage members of the LSC to test to at least 67 markers. It not only gives the best proof of memembership, but also greatly enhances comparison to other men in the cluster.
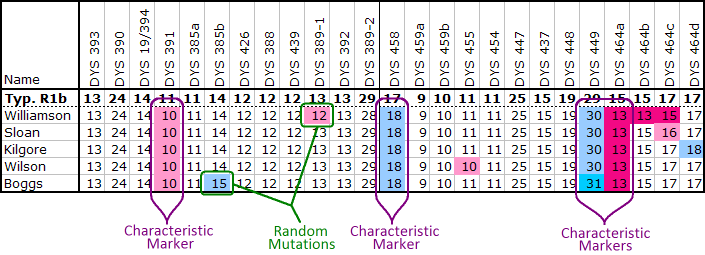
Of course not all group members will have all of these characteristic values. Mutations can happen to any marker, even the characteristic ones. Thus each member will also have markers whose values differ from both the typical R1b values and the characteristic values. (See ySearch ID 55GU9 for the R1b modal value.)
One could also look at genetic distance from the LSC modal haplotype. Such a comparison could be done on y-Search. Typically, most LSC men are withing a distance of 12 at 67-markers from the LSC modal. In terms of genetic distance (count of mutations between the haplotypes), that depends very much on the number of markers compared. It is not sufficient that the candidate's sequence be within a certain 'genetic distance' of another cluster member. Instead, the candidate's haplotype should be consistent with what we would expect of a descendant of the cluster's founder. In particular, any "perculiarities" in the founder's haplotype we would expect to find in the candidate's haplotype.
